Package randomForest has importance() to estimate the importance of variables.
The example in the reference manual has this:
> library(randomForest)
> data(mtcars)
> head(mtcars)
mpg cyl disp hp drat wt qsec vs am gear carb
Mazda RX4 21.0 6 160 110 3.90 2.620 16.46 0 1 4 4
Mazda RX4 Wag 21.0 6 160 110 3.90 2.875 17.02 0 1 4 4
Datsun 710 22.8 4 108 93 3.85 2.320 18.61 1 1 4 1
Hornet 4 Drive 21.4 6 258 110 3.08 3.215 19.44 1 0 3 1
Hornet Sportabout 18.7 8 360 175 3.15 3.440 17.02 0 0 3 2
Valiant 18.1 6 225 105 2.76 3.460 20.22 1 0 3 1
> mtcars.rf <- randomForest(mpg ~ ., data=mtcars, ntree=1000, keep.forest=FALSE, importance=TRUE)
> importance(mtcars.rf)
%IncMSE IncNodePurity
cyl 16.050788 171.09822
disp 18.868236 232.56372
hp 17.031602 198.29501
drat 7.728328 64.23068
wt 18.595598 260.77604
qsec 5.607246 33.88488
vs 5.124934 26.49292
am 3.938463 13.72707
gear 4.482608 18.85271
carb 7.823431 33.94279
> importance(mtcars.rf, type=1)
%IncMSE
cyl 16.050788
disp 18.868236
hp 17.031602
drat 7.728328
wt 18.595598
qsec 5.607246
vs 5.124934
am 3.938463
gear 4.482608
carb 7.823431
In importance(), type=1 shows mean squared error increase if each variable is removed from the predictors. Type 2 shows increase in node impurity averaged over all trees.
To visualize:
> varImpPlot(mtcars.rf)
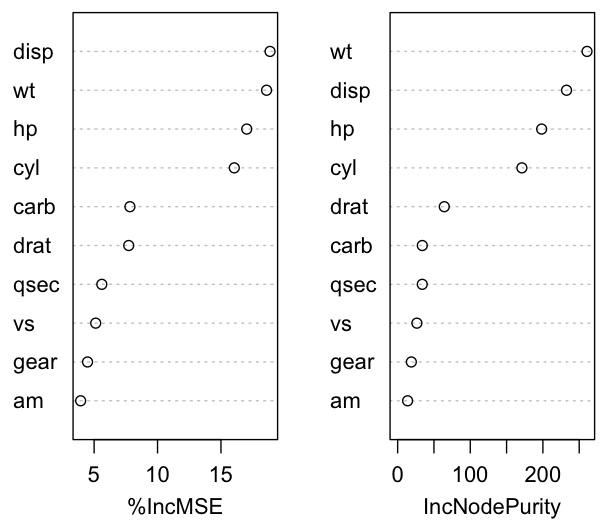
To get the top three important variables:
> mtcars.imp <- importance(mtcars.rf, type=1)
> mtcars.imp[order(mtcars.imp, decreasing=TRUE),]
disp wt hp cyl carb drat qsec vs
18.868236 18.595598 17.031602 16.050788 7.823431 7.728328 5.607246 5.124934
gear am
4.482608 3.938463
> names(mtcars.imp[order(mtcars.imp, decreasing=TRUE),])[1:3]
[1] "disp" "wt" "hp"
Thus we get disp, wt, and hp.