> a = aov(iris$Petal.Width ~ iris$Species)
> a
Call:
aov(formula = iris$Petal.Width ~ iris$Species)
Terms:
iris$Species Residuals
Sum of Squares 80.41333 6.15660
Deg. of Freedom 2 147
Residual standard error: 0.20465
Estimated effects may be unbalanced
Look. You should call summary on the ret. val of aov to get the following statistics.
> summary(a)
Df Sum Sq Mean Sq F value Pr(>F)
iris$Species 2 80.413 40.207 960.01 < 2.2e-16 ***
Residuals 147 6.157 0.042
—
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Because P value is very small, H0 is rejected; Petal.Width is different depending on Species.
We can draw a boxplot to visualize this:
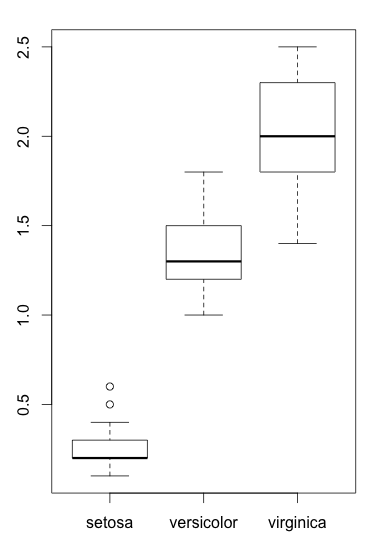
Some texts[1] explain that F statistics in ANOVA is (between class variance) / (within class variance) while others say F is (treatment) / (random error), but they basically evaluate the same thing.
ANOVA assumes a couple of things.
a) variance should be the same across classes.
b) error should be independent, and gaussian.
c) data from each class is independent.
In the above, I didn’t do much to show that the assumption holds, but one can run
plot(a)
to get some graphs like ‘residual vs fitted’, ‘qq plot’, etc.
References)
1. http://homepages.inf.ed.ac.uk/rbf/CVonline/LOCAL_COPIES/SHUTLER2/node1.html